This is an old revision of the document!
BLAST
 In bioinformatics, BLAST is an algorithm and program for comparing primary biological sequence information, such as the amino-acid sequences of proteins or the nucleotides of DNA and/or RNA sequences.
In bioinformatics, BLAST is an algorithm and program for comparing primary biological sequence information, such as the amino-acid sequences of proteins or the nucleotides of DNA and/or RNA sequences.
Basic Local Alignment Search Tool
BLAST finds regions of similarity between biological sequences. The program compares nucleotide or protein sequences to sequence databases and calculates the statistical significance. 1)
BLAST is the NCBI/NIH (aka US government) repository for genomic and proteomic sequences, amongst other things. It is where all genome scientists around the world deposit their sequences if they make a discovery. Its main function is to allow comparison of gene sequences and discovery of sequences that match one that you might have come across in your experiment.
What’s a gene sequence? It’s a line of code, made up of any combination of 4 letters in a sequence. BLAST has two sections - nucleotide (BLASTn) and protein (BLASTp). BLASTp deals with amino acid sequences, in just the same way as nucleotide sequences.
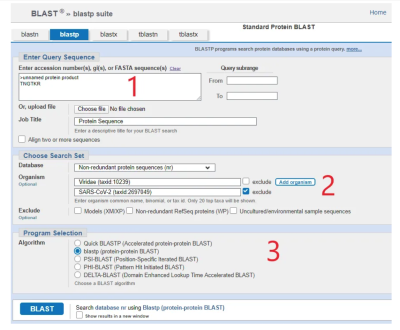
How to BLAST your way to the truth about the origins of COVID-19
Using BLAST is easy. I'm going to show you how easy and how to prove that SARS-Cov-2 is man-made Dr Ah Kahn Syed - Dec 28, 2021 2)
